We are devoted to developing advanced lineage tracing tools and applying them to solve important biological questions. Please check out our publications for our latest works, including
- CoSpar, a fate-prediction method based on state and lineage information (Nature Biotechnology, 2022)
- DARLIN, a state-of-the-art lineage tracing mouse model with enormous barcoding capacity (Cell, 2023)
- Camellia-seq, the first single-cell multi-omic lineage profiling method (Cell, 2023).
- MethylTree, the first non-invasive lineage tracing method based on DNA methylation epimutations (Nature Methods, 2025).
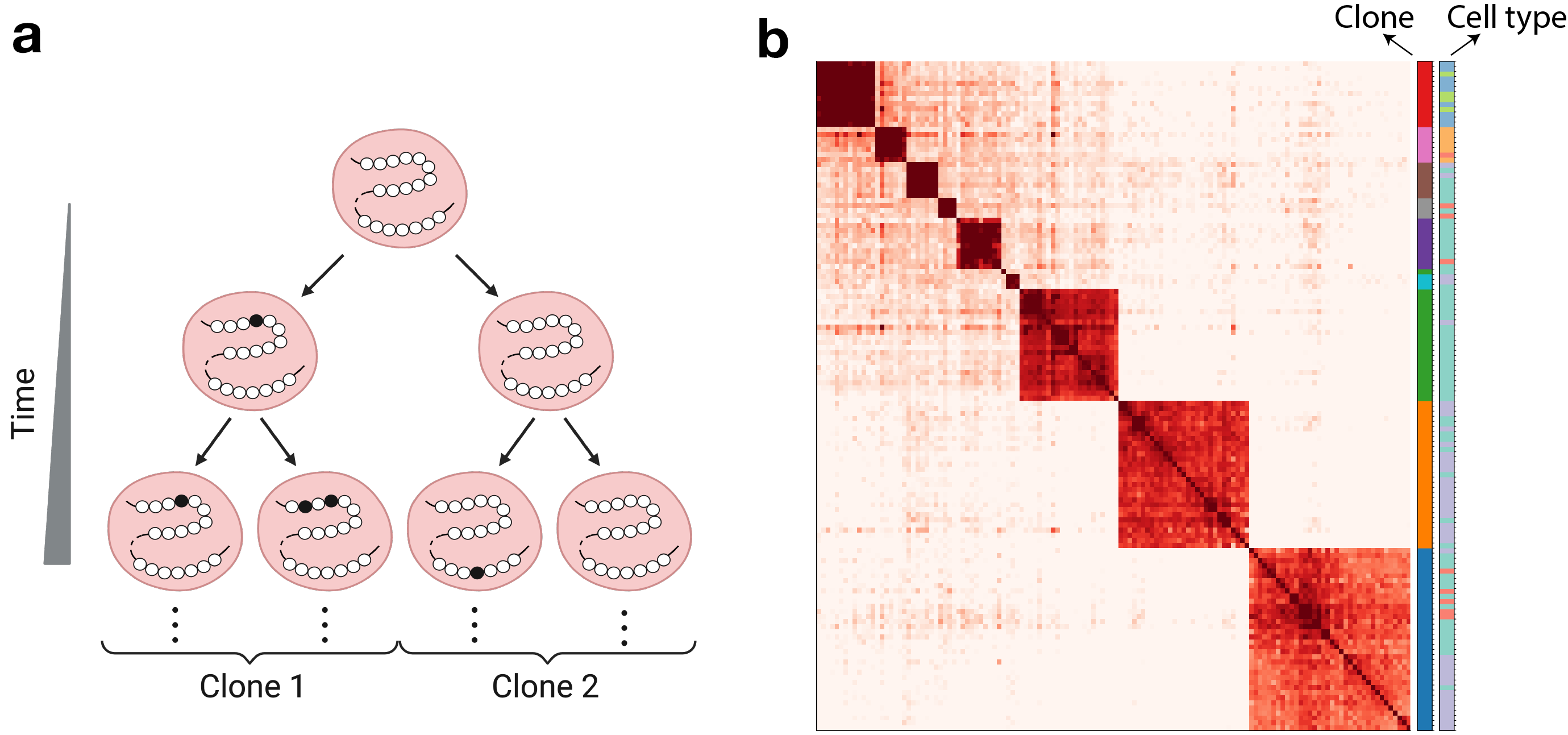
MethylTree opens the door for lineage tracing in humans. Currently, our whole lab is revolved around DNA methylation. We have established a reliable platform to jointly generate DNA methylome and transcriptome in single cells. Half of the lab members are devoted to platform development and data generation, while the remaining are working on computational analysis and algorithm development. These efforts are integrated together towards the same goal: resovling cell lineages in humans. Below are our research directions:
- We are now collaborating with hospitals to collect clinical samples and carry out non-invasive single-cell multiomic lineage tracing on humans.
- We are also seeking to further improve methylation-based lineage tracing approach, both experimentally and computationally.
- We are also carrying out experiments to further our understanding of DNA methylation biology.
If you also share our passion that DNA methylation is a powerful source of information for lineage tracing, please join our team!

